Synthetic control of yeast culture growth rates (ongoing)
I am finishing a project started as a postdoc in the Klavins Lab at UW with Orlando de Lange and Alberto Carignano that we are about to publish. We engineered yeast to induce growth arrest using a number of genes/inducers that we then characterized. We then used it in a cool application. I work both …
Design of Languages for Systems and Synthetic Biology
Context We propose to encompass context-dependent genomic information and mathematical models in a logical and structured fashion through the use of attribute grammars. Attribute Grammars (AG) provide means to compute a mathematical model (possibly encoding phenotypic traits) from a genetic construct. AGs are a framework for biological Domain Specifi c Languages (DSLs) that, unlike current formalisms …
Research on Language and Biology
The theory: Formal language and Genetic This is related to the work I have done at Peccoud’s synthetic biology lab from 2008 to 2013: Formal languages and DNA molecules Design of Languages for Systems and Synthetic Biology Biological Languages to Design Natural Genomes [Slides] Formal languages to map Genotype to Phenotype in Natural Genomes. Application: …
PACHA.YEAST: Morphogenetic engineering of yeast Microcolonies
Incas regarded space and time as a single concept, referred to as pacha ( Quechua: pacha, Aymara: pacha). A poster of my work on PACHA.YEAST: Highlights: Lab automation with Apprent’yeast: learned patterns to automatically design plasmids from lab data (inferred a probabilistic context-free grammar (PCFG)) Adam, L. & Klavins, E. (2015). archiYEAST: a command-line synthetic yeast …
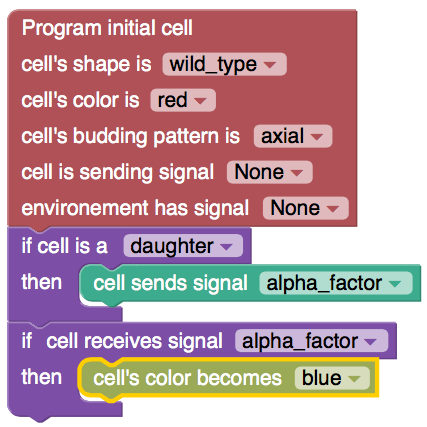
LYSD: Language for Yeast Spatiotemporal Development
LYSD is a language a created to specify a phenotype. It can be compiled down into a genetic designs, which ultimately can be automatically build into a lab (check out Aquarium from the Klavins lab; I used it with a python API). We could formally verify a programmed phenotype: microscopy images can be analysed and …
Gro for snowflake yeast
Gro is a software written in C++ using QT. Gro is a programming language and a 2D simulation environment for cells. The physics and front end has been implemented for e.coli. Specification and simulation of synthetic multicelled behaviors. Jang SS(1), Oishi KT, Egbert RG, Klavins E. Author information: (1)Department of Electrical Engineering, University of Washington, …
The Synthetic Biology Open Language (SBOL) provides a community standard for communicating designs in synthetic biology
Authors: Michal Galdzicki, Kevin P Clancy, Ernst Oberortner, Matthew Pocock, Jacqueline Y Quinn, Cesar A Rodriguez, Nicholas Roehner, Mandy L Wilson, Laura Adam, J Christopher Anderson, Bryan A Bartley, Jacob Beal, Deepak Chandran, Joanna Chen, Douglas Densmore, Drew Endy, Raik Grünberg, Jennifer Hallinan, Nathan J Hillson, Jeffrey D Johnson, Allan Kuchinsky, Matthew Lux, Goksel Misirli, …
Development of a domain-specific genetic language to design Chlamydomonas reinhardtii expression vectors
Authors Mandy L Wilson, Sakiko Okumoto, Laura Adam, Jean Peccoud Publication date 2013/11/8 Journal Bioinformatics Volume30 Issue2 Pages 251-257 Abstract Motivation: Expression vectors used in different biotechnology applications are designed with domain-specific rules. For instance, promoters, origins of replication or homologous recombination sites are host-specific. Similarly, chromosomal integration or viral delivery of an expression cassette imposes specific …
[Slides DEFENSE] Mapping Genotype to Phenotype using Attribute Grammar
MAPPING GENOTYPE TO PHENOTYPE USING ATTRIBUTE GRAMMAR Doctoral Dissertation Defense Thursday, July 25th, 2013 – 1:00PM Virginia Bioinformatics Institute – Room 325LAURA ADAM Genetics, Bioinformatics and Computational Biology Program ADVISORY COMMITTEE: Jean Peccoud (Chair) David Bevan, Harold Garner, François Képès, Naren Ramakrishnan, John Tyson Abstract Over the past 10 years, several synthetic biology research groups …
[Slides] Formal languages to map Genotype to Phenotype in Natural Genomes.
Adam, L. Formal languages to map Genotype to Phenotype in Natural Genomes. GBCB seminar, 2012. Speaker: Laura Adam, GBCB Doctoral Candidate Advisor: Jean Peccoud, VBI Title: Formal languages to map Genotype to Phenotype in Natural Genomes Abstract: To formalize Genotype-to-Phenotype mapping and confer predictive powers, missing in current approaches, information from genomic databases and mathematical …