Low-cost, modular, multi-strain diagnostic (ongoing)
This is the application part of the project I am finishing. We engineered yeast to induce growth arrest using a number of genes/inducers that we then characterized. We then used them as a diagnostic tool. We used nonlinear differential equations. We model our co-culture experiments and made predictions. We used a flow cytometer to collect …
Synthetic control of yeast culture growth rates (ongoing)
I am finishing a project started as a postdoc in the Klavins Lab at UW with Orlando de Lange and Alberto Carignano that we are about to publish. We engineered yeast to induce growth arrest using a number of genes/inducers that we then characterized. We then used it in a cool application. I work both …
Design of Languages for Systems and Synthetic Biology
Context We propose to encompass context-dependent genomic information and mathematical models in a logical and structured fashion through the use of attribute grammars. Attribute Grammars (AG) provide means to compute a mathematical model (possibly encoding phenotypic traits) from a genetic construct. AGs are a framework for biological Domain Specifi c Languages (DSLs) that, unlike current formalisms …
Research on Language and Biology
The theory: Formal language and Genetic This is related to the work I have done at Peccoud’s synthetic biology lab from 2008 to 2013: Formal languages and DNA molecules Design of Languages for Systems and Synthetic Biology Biological Languages to Design Natural Genomes [Slides] Formal languages to map Genotype to Phenotype in Natural Genomes. Application: …
ebio·ome: the DNA space explorer.
Talk at SynBioBeta 2017 http://sf2017.synbiobeta.com/speakers/laura-adam/
PACHA.YEAST: Morphogenetic engineering of yeast Microcolonies
Incas regarded space and time as a single concept, referred to as pacha ( Quechua: pacha, Aymara: pacha). A poster of my work on PACHA.YEAST: Highlights: Lab automation with Apprent’yeast: learned patterns to automatically design plasmids from lab data (inferred a probabilistic context-free grammar (PCFG)) Adam, L. & Klavins, E. (2015). archiYEAST: a command-line synthetic yeast …
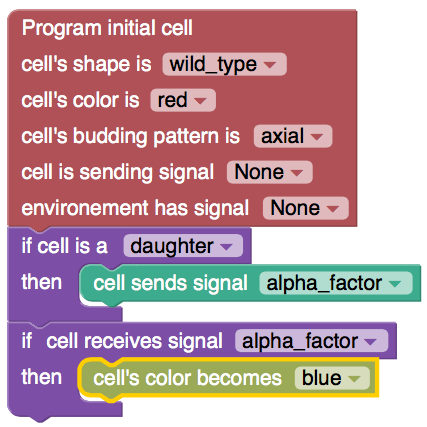
LYSD: Language for Yeast Spatiotemporal Development
LYSD is a language a created to specify a phenotype. It can be compiled down into a genetic designs, which ultimately can be automatically build into a lab (check out Aquarium from the Klavins lab; I used it with a python API). We could formally verify a programmed phenotype: microscopy images can be analysed and …
Judging at iGEM
2010-2016: Judge at the Jamboree 2014: Committee member Policy and Practices New track 2012: Judge at aGEM, Edmonton, Alberta, Canada (invited) 2010-2011: Advisor Virginia Tech Team and Virginia Tech-ENSIMAG Biosecurity Team
[OVERVIEW] My BioCAD tools
Design: GenoCAD – Rule-based genetic design tools user-friendly and Domain Specific Language M. Galdzicki, K. P. Clancy, E. Oberortner, M. Pocock, J. Y. Quinn, C. a Rodriguez, N. Roehner, M. L. Wilson, L. Adam, J. C. Anderson, B. a Bartley, J. Beal, D. Chandran, J. Chen, D. Densmore, D. Endy, R. Grünberg, J. Hallinan, N. …