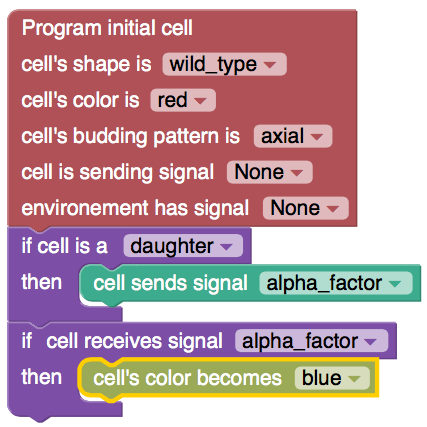
LYSD: Language for Yeast Spatiotemporal Development
LYSD is a language a created to specify a phenotype.
It can be compiled down into a genetic designs, which ultimately can be automatically build into a lab (check out Aquarium from the Klavins lab; I used it with a python API).
We could formally verify a programmed phenotype: microscopy images can be analysed and parsed against a LYSD program.
It can be simulated using Gro.
Formally, LYSD is based on attribute graph grammars.
I also used blockly (similar to Scratch) to let user program yeast microcolony in LYSD.
